Diverse, abundant microbial communities are found across all of the earth’s ecosystems, from the human intestinal tract to deep water hydrothermal vents to rainforest canopies. Scientists are interested in how these different species of bacteria interact and what that could mean for advancements in science and medicine.
Biofilms and Bacterial Colonies
Both in and outside of labs, bacteria frequently form a biofilm when attaching themselves to surfaces. A biofilm contains microbial cells and an extracellular polymeric substance (EPS) matrix; this EPS has a defined architecture, and it allows cells to exchange genetic material.
Within a biofilm, bacteria have a range of social interactions that include competition, predation, and cooperation. Using secondary metabolites — natural compounds involved in the organism’s homeostasis — bacteria can signal, sense, and respond to other bacteria in what’s called cell-cell communication.
Bacterial communities can also be shaped by mechanical forces; in dense colonies, bacteria push against each other like commuters on a crowded subway platform. The mechanical forces that shape multispecies colonies, however, have not been widely studied.
Study Background and Expectations
Due to the lack of research on this topic, a group of scientists from University of California San Diego chose to study the mechanical interactions between two very different species of bacteria — Acinetobacter baylyi and Escherichia coli —in flower-like patterns in multispecies bacterial colonies. A. baylyi is a gram-negative bacteria that moves easily on soft surfaces through a twitching motility and possesses a secretion system that allows it to kill other bacteria on contact. E. coli is a rod-shaped gram-negative bacteria that is almost non-motile on soft surfaces.
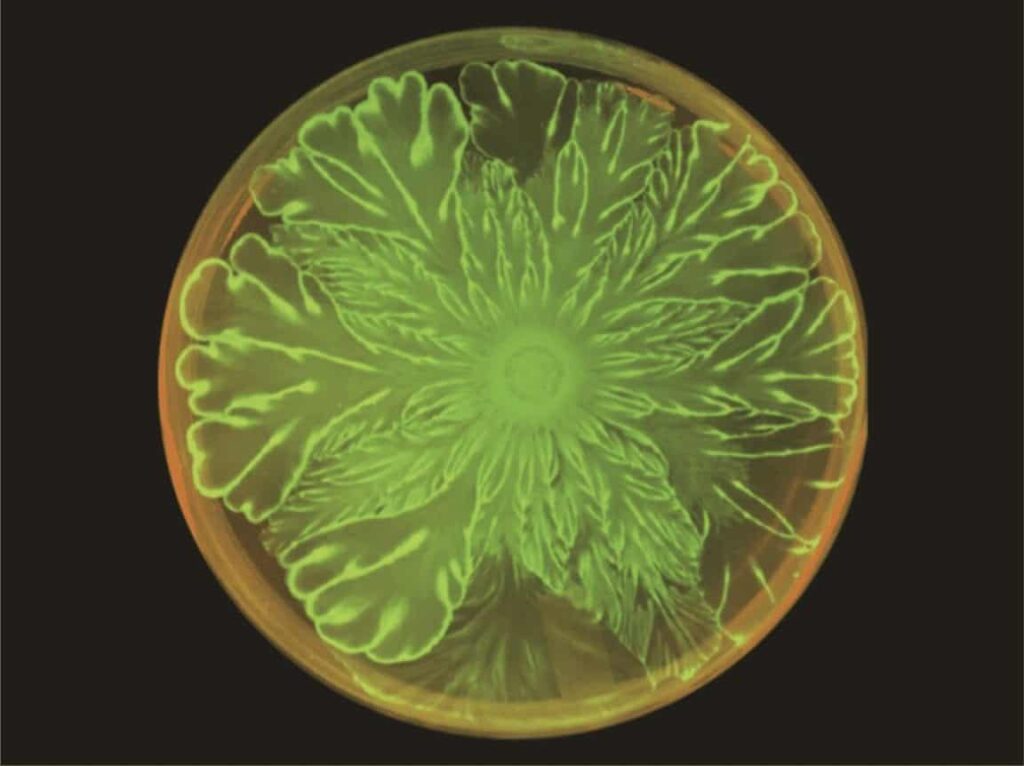
The scientists expected large differences in mechanical stress distribution, and thus complex formations in the colony’s spatiotemporal dynamics. When mixed together in a colony, these two species produced intricate flower patterns not shown when either species was grown by itself in a control. The study focused on whether biological cell-cell communication or mechanical interaction guided the pattern formation.
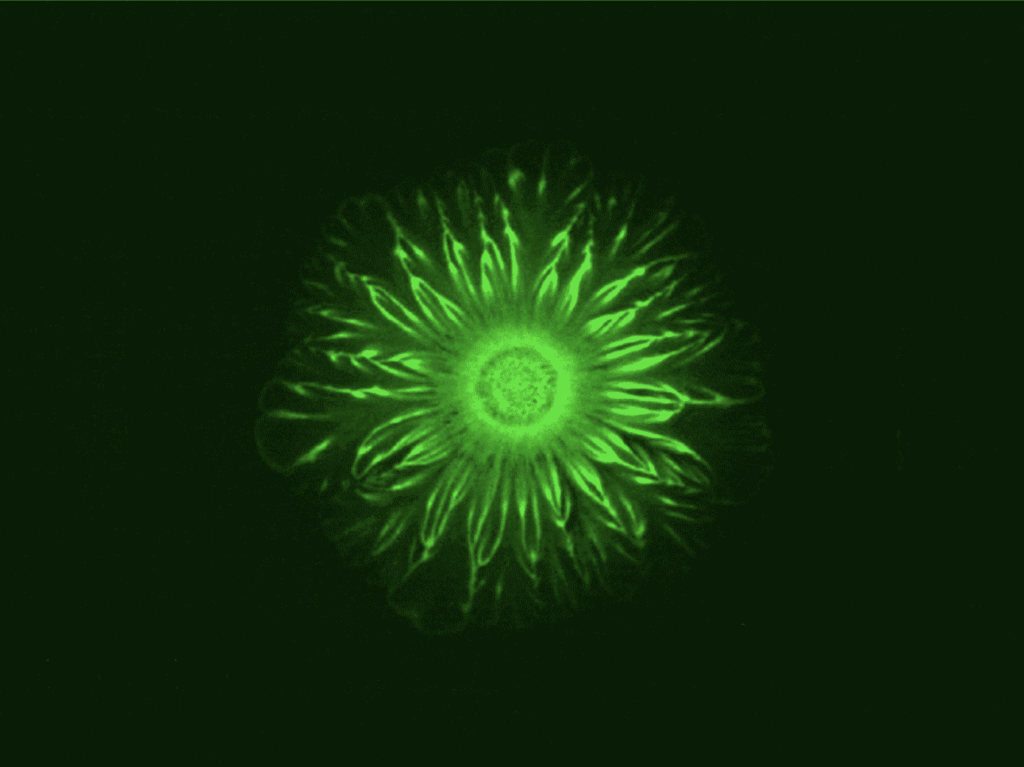
The Experiment
The experiment used a 10:1 ratio of E. coli and A. baylyi in soft agar that was grown at 37 degrees Celsius for three days. Mutations of A. baylyi that lacked the ability to kill E. coli on contact were used to rule out that possible causality. A second control experiment used mobility-impaired mutations of A. baylyi and did not result in the flower pattern formation. Thus, the patterns could be attributed to the mechanical interactions between the two bacterial strains — not the cell-cell communication.
After eight hours, the colony front began to visibly undulate. The fact that both species grew and spread rapidly — not just the mobile A. baylyi — implies the E. coli is being pushed in front of the A. baylyi, with the A. baylyi acting like a snowplow. The E. coli cells accumulate at the expanding boundary of the bacterial colony, making the boundary unstable and producing the undulating flower patterns.
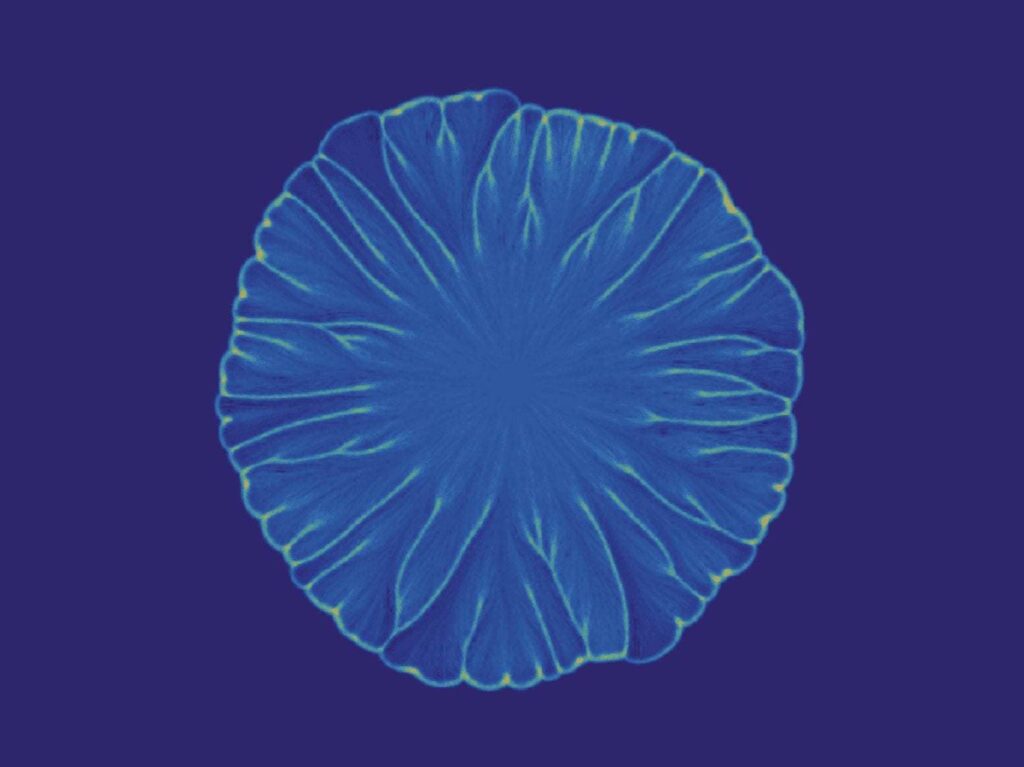
Real-Life Implications
Beyond the simple beauty found under the microscope while watching some of our planet’s tiniest microorganisms, this experiment has implications for the study of biofilms. Biofilms have been shown to play a key role in chronic infections — such as airway infections in cystic fibrosis and COPD, endocarditis, recurrent sinusitis, and poorly healing ulcers — in human hosts. Understanding how mixed colonies of bacteria interact within a biofilm will assist scientists and medical professionals in developing treatments for patients with chronic infections.
Since 1995, QPS has provided discovery, preclinical, and clinical drug development services. An award-winning leader focused on bioanalytics and clinical trials, QPS is known for proven quality standards, technical expertise, a flexible approach to research, client satisfaction and turnkey laboratories and facilities. For more information, visit www.qps.com or email info@qps.com.







